Hand on on exploration results
Result fusion files
Exploration Fusion results
====
Data Exploration
In the following section we will use the ucsc genome browser’s online blat to explore a number of example positive and negative fusion transcripts. This part is based on tutorial created by Andrew McPherson The aim of this part is to understand the procedure to validate a fusion transcript
Hand On 1
Now we can take a look of Breast cancer cell line. We start to analyze starfusion results in detail. Move on the STAR-FUSION directry of HCC1395 directory.
cd ~/fusion/tutorial-fusion-transcript-2017/Results/HCC1395
then uncompress the directory:
tar xvfz starfusion.tar.gz
check the directory:
ls
defuse.tar.gz starfusion starfusion.tar.gz
move on the star fusion directory:
cd starfusion/
Now look the starfusion.fusion_candidates.txt and serarch some fusions:
FROM: High-throughput detection of fusion genes in cancer using the Sequenom MassARRAY platform
Maryou BK Lambros, Paul M Wilkerson, Rachael Natrajan, Neill Patani, Vidya Pawar, Radost Vatcheva, Marthe Mansour, Mirja Laschet, Beatrice Oelze, Nicholas Orr, Susanne Muller and Jorge S Reis-Filho
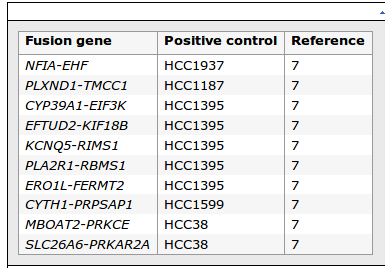
Please check ERO1L-FERMT2 and CYP39A1-EIF3K
**(root) um49@m49:~/fusion/tutorial-fusion-transcript-2017/Results/HCC1395/starfusion$ cat starfusion.fusion_candidates.txt |grep CYP39A1|grep EIF3K**
EIF3K--CYP39A1 170 109 EIF3K^ENSG00000178982.5 chr19:39123318:+ 0CYP39A1^ENSG00000146233.3 chr6:46607405:- 0
**(root) um49@m49:~/fusion/tutorial-fusion-transcript-2017/Results/HCC1395/starfusion$ cat starfusion.fusion_candidates.txt |grep ERO1L|grep FERMT2**
ERO1L--FERMT2 3 2 ERO1L^ENSG00000197930.8 chr14:53138348:- 0FERMT2^ENSG00000073712.9 chr14:53348187:- 0
Use cat and grep for search the other fusions:
cat starfusion.fusion_candidates.txt |grep ####|grep ######
Hand On 2
Go to Defuse results and see the file type. Defuse are perl tool very powerfull create by Andrew McPherson. DeFuse is a software package for gene fusion discovery using RNA-Seq data. The software uses clusters of discordant paired end alignments to inform a split read alignment analysis for finding fusion boundaries. The software also employs a number of heuristic filters in an attempt to reduce the number of false positives and produces a fully annotated output for each predicted fusion.
Move on the STAR-FUSION directry of HCC1395 directory.
cd ~/fusion/tutorial-fusion-transcript-2017/Results/HCC1395
then uncompress the directory:
tar xvf defuse.tar.gz
check the directory:
**(root) um49@m49:~/fusion/tutorial-fusion-transcript-2017/Results/HCC1395$ ls**
defuse defuse.tar.gz starfusion starfusion.tar.gz
cd defuse/
The information how to interpeter the output are describe in this page
Please go to defuse results and extract the sequnce from PLA2R1-RBMS1.
cat results.filtered.tsv |grep PLA2R1
The following two sequence predicted by defuse represent two distinct splice variants of the PLA2R1-RBMS1 fusion.
>771050
AAGATGCAAGAAACTGTGCTGTTTATAAGGCAAACAAAACATTGCTGCCCTTACACTG
TGGTTCCAAACGTGAATGGATATGCAAAATCCCAAGAGATGTGAAACCCAAGATTCCG
TTCTGGTACCAGTACGATGTACCCTGGCTCTTTTATCAGGATGCAGAATACCTTTTTC
ATACCTTTGCCTCAGAATGGTTGAACTTTGAGTTTGTCTGTAGCTGGCTGCACAGTGA
TCTTCTCACAATTCATTCTGCACATGAGCAAGAATTCATCCACAGCAAAATAAAAGCG
|CAACAGGAACAAGATCCTACCAACCTCTACATTTCTAATTTGCCACTCTCCATGGAT
GAGCAAGAACTAGAAAATATGCTCAAACCATTTGGACAAGTTATTTCTACAAGGATAC
TACGTGATTCCAGTGGTACAAGTCGTGGTGTTGGCTTTGCTAGGATGGAATCAACAGA
AAAATGTGAAGCTGTTATTGGTCATTTTAATGGAAAATTTATTAAGACACCACCAGGA
GTTTCTGCCCCCACAGA
>770399
GGATTGGATTTAATAAAAGAAACCCACTGAATGCCGGCTCATGGGAGTGGTCTGATAG
AACTCCTGTTGTCTCTTCGTTTTTAGACAACACTTATTTTGGAGAAGATGCAAGAAAC
TGTGCTGTTTATAAGGCAAACAAAACATTGCTGCCCTTACACTGTGGTTCCAAACGTG
AATGGATATGCAAAATCCCAAGAGATGTGAAACCCAAGATTCCGTTCTGGTACCAGTA
CGATGTACCCTGGCTCTTTTATCAGGATGCAGAATACCTTTTTCATACCTTTGCCTCA
GAATGGTTGAACTTTGAGTTTGTCTGTAGCTGGCTGCACAGTGATCTTCTCACAATTC
ATTCTGCACATGAGCAAGAATTCATCCACAGCAAAATAAAAGC|GCAACAGGAACAAG
ATCCTACCAACCTCTACATTTCTAATTTGCCACTCTCCATGGATGAGCAAGAACTAGA
AAATATGCTCAAACCATTTGGACAAGTTATTTCTACAAGGATACTACGTGATTCCAGT
GGTACAAGTCGTGGTGTTGGCTTTGCTAGATTTTTTTTTTTTTTTGTGAGAAATA
Web-blatting link them will give you the following list of alignments. Please remember Defuse use hg19.
ACTIONS QUERY SCORE START END QSIZE IDENTITY CHRO STRAND START END SPAN
---------------------------------------------------------------------------------------------------
browser details 770399 389 1 392 576 100.0% 2 - 160832579 160836405 3827
browser details 770399 184 392 576 576 100.0% 2 - 161159310 161159999 690
browser details 770399 155 392 550 576 98.8% 12 - 66628514 66628672 159
browser details 770399 78 392 547 576 75.0% 12 - 94818206 94818361 156
browser details 770399 27 361 391 576 96.6% 2 - 5782745 5783131 387
browser details 770399 22 104 125 576 100.0% 5 + 148444986 148445007 22
browser details 770399 21 438 458 576 100.0% 6 - 158134531 158134551 21
browser details 770399 20 265 284 576 100.0% 5 - 84000186 84000205 20
browser details 770399 20 235 266 576 81.3% 2 + 74143821 74143852 32
browser details 771050 288 1 290 538 100.0% 2 - 160832579 160833890 1312
browser details 771050 241 290 538 538 98.4% 12 - 66628424 66628672 249
browser details 771050 238 290 528 538 100.0% 2 - 161157162 161159999 2838
browser details 771050 83 290 453 538 75.5% 12 + 56965481 56965649 169
browser details 771050 39 462 520 538 83.1% 3 + 29804414 29804472 59
browser details 771050 27 259 289 538 96.6% 2 - 5782745 5783131 387
browser details 771050 27 441 472 538 96.7% 3 + 192743568 192743603 36
browser details 771050 25 461 493 538 76.7% 2 - 182283137 182283166 30
browser details 771050 22 2 23 538 100.0% 5 + 148444986 148445007 22
browser details 771050 21 336 356 538 100.0% 6 - 158134531 158134551 21
browser details 771050 20 163 182 538 100.0% 5 - 84000186 84000205 20
browser details 771050 20 1 24 538 91.7% 1 + 162028403 162028426 24
Click ‘browser’ for the first alignment for the alignment to the PLA2R1 gene.
Do you expect the genomic breakpoint to be upstream (on the left) or downstream (on the right) relative to the aligned sequences?
Go back and click ‘browser’ for the second alignment for the alignment to the RBMS1 gene.
Do you expect the genomic breakpoint to be upstream (on the left) or downstream (on the right) relative to the aligned sequences?
The following sequence is a destruct breakpoint prediction most likely associated with the fusion.
>destruct_31240
TGCAAAAGATCTGGAAAAATGCAGTCTGGTATTTACACATAATTTAAGTTCACAGTGC
AACTGCTCCCATAACCCTAGCTGAAACTGTCTCTTCTTAGTCATTTTTAATTTTCCAA
GATAACTTGGCAAAGCTATTGTTGTTGACATAATAAAGACTGGGCAGAAGGCTTACCT
AGCAAAGCCAACACCACGACTTGTACCACTGGAATCACGTAGTATCCTTGTAGAAATA
ACTTGTCCAAATGGTTTGAGCATATTTTCTAGTTCTTGCTCATCCATGGAGAGTGGCA
AATTAGAAATGTAGAGGTTGGTAGGATCTTGTTCCTGTTGCTAAAACAGAAGAGAGTG
TTGTCCATTAATTTCCAACAGAAGGTGAGATATTTATGTTAACACACCTATTTTTATT
AGCTACTTTCTTTGCTCAAGTCCTTTTAAAGTACTCAGAACCTCAGAACACCAAAGTC
ACCCTGGACTCTTGAAAATAGTGTCTGAAGCTTGGACAA[AA]AAAAAGTAATATTAG
AAAATGAATTCATTTTCTGACAAAAAATTATTGGCTCATCCTCTCAGTTATTTACCCT
CTCAGTGATTTATAATTCATTGCATATGTCACATGTATTTGAAAAACAATTCAAGGTA
TCAAGGCATCATTAGTATAAAGATACTGATTTTAGGTATTAGTCTGATTGCTAAGCTT
TAAGCAGTATAAGCTTTCCTTCCCATTCAAATAGAGAGACACAATATAGGACAAAAGA
ATACTACAGAGTGCCCAGTGTTTGACAACTAGAAAATTATCCTTTTGATGAGTTCATG
TCCTTTGCAGGGACATGGATGAAGCTGGAAACCATCAATCTCAGCAAACTAACACATG
AACAGAAAACCAAACACCGCATGTTCTCACTCATAAGTGGGAGCTGAACAATGAGAAC
ACATGGACACAGGGAGGGGAACATCACACACTGGGGCCTGTCGG
Add the breakpoint sequence to the web blat query, click on the same alignments as above, and zoom out to see the position of the breakpoint relative to the fusion.
Hand On 3
Now we want to evaluete the result obtained from JAFFA on the simulated data. Open jaffa result.csv and extract the sequencing and blat the results.
**cd ~/fusion/tutorial-fusion-transcript-2017/Results/JAFFA/Simulazione_50x/**
**(root) um49@m49:~/fusion/tutorial-fusion-transcript-2017/Results/JAFFA/Simulazione_50x$ cat jaffa_results.csv |grep ETV6**
"reads_50x","ETV6:NTRK3","chr12",11869968,"+","chr15",87940752,"-",Inf,"169",19,NA,FALSE,TRUE,"Locus_1_Transcript_77/274_Confidence_0.022_Length_8978",1279,"LowConfidence","Yes"
Now extract thename of the contig of ETV6-NTRK3 for search on fasta file:
**(root) um49@m49:~/fusion/tutorial-fusion-transcript-2017/Results/JAFFA/Simulazione_50x$ cat jaffa_results.csv |grep ETV6|awk -F"," '{print $15}'**
"Locus_1_Transcript_77/274_Confidence_0.022_Length_8978"
On .fasta exxtract that sequence:
(root) um49@m49:~/fusion/tutorial-fusion-transcript-2017/Results/JAFFA/Simulazione_50x$ cat jaffa_results.fasta |grep -A1 Locus_1_Transcript_77/274_Confidence_0.022_Length_8978
>reads_50x---ETV6:NTRK3---Locus_1_Transcript_77/274_Confidence_0.022_Length_8978
GTCCCGGGTCCCCGCGCCGCGCCGCGACCTGCAGACCCCGCCGCCGCGCTCGGGCCCGTCTCCCACGCCCCCGCCGCCCCGCGCGCCCAACTCCGCCGGCCGCCCCGCCCCGCCCCGCGCGCTCCAGACCCCCGGGGCGGCTGCCGGGAGAGATGCTGGAAGAAACTTCTTAAATGACCGCGTCTGGCTGGCCG.......
This is the complet sequence:
>reads_50x---ETV6:NTRK3---Locus_1_Transcript_77/274_Confidence_0.022_Length_8978
GTCCCGGGTCCCCGCGCCGCGCCGCGACCTGCAGACCCCGCCGCCGCGCTCGGGCCCGTCTCCCACGCCCCCGCCGCCCCGCGCGCCCAACTCCGCCGGCCGCCCCGCCCCGCCCCGCGCGCTCCAGACCCCCGGGGCGGCTGCCGGGAGAGATGCTGGAAGAAACTTCTTAAATGACCGCGTCTGGCTGGCCGTGGAGCCTTTCTGGGTTGGGGAGAGGAAAGGAAAGTGGAAAAAACCTGAGAACTTCCTGATCTCTCTCGCTGTGAGACATGTCTGAGACTCCTGCTCAGTGTAGCATTAAGCAGGAACGAATTTCATATACACCTCCAGAGAGCCCAGTGCCGAGTTACGCTTCCTCGACGCCACTTCATGTTCCAGTGCCTCGAGCGCTCAGGATGGAGGAAGACTCGATCCGCCTGCCTGCGCACCTGCGCTTGCAGCCAATTTACTGGAGCAGGGATGACGTAGCCCAGTGGCTCAAGTGGGCTGAAAATGAGTTTTCTTTAAGCCAATTGACAGCAACACGTTTGAAATGAATGGCAAAGCTCTCCTGCTGCTGACCAAAGAGGACTTTCGCTATCGATCTCCTCATTCAGGTGATGTGCTCTATGAACTCCTTCAGCATATTCTGAAGCAGAGGAAACCTCGGATTCTTTTTTCACCATTCTTCCACCCTGGAAACTCTATACACACACAGCCGGAGGTCATACTGCATCAGAACCATGAAGAAGATAACTGTGTCCAGAGGACCCCCAGGCCATCCGTGGATAATGTGCACCATAACCCTCCCACCATTGAACTGTTGCACCGCTCCAGGTCACCTATCACGACAAATCACCGGCCTTCTCCTGACCCCGAGCAGCGGCCCCTCCGGTCCCCCCTGGACAACATGATCCGCCGCCTCTCCCCGGCTGAGAGAGCTCAGGGACCCAGGCCGCACCAGGAGAACAACCACCAGGAGTCCTACCCTCTGTCAGTGTCTCCCATGGAGAATAATCACTGCCCAGCGTCCTCCGAGTCCCACCCGAAGCCATCCAGCCCCCGGCAGGAGAGCACACGCGTGATCCAGCTGATGCCCAGCCCCATCATGCACCCTCTGATCCTGAACCCCCGGCACTCCGTGGATTTCAAACAGTCCAGGCTCTCCGAGGACGGGCTGCATAGGGAAGGGAAGCCCATCAACCTCTCTCATCGGGAAGACCTGGCTTACATGAACCACATCATGGTCTCTGTCTCCCCGCCTGAAGAGCACGCCATGCCCATTGGGAGAATAGCagTGTGCAGCACATTAAGAGGAGAGACATCGTGCTGAAGCGAGAACTGGGTGAGGGAGCCTTTGGAAAGGTCTTCCTGGCCGAGTGCTACAACCTCAGCCCGACCAAGGACAAGATGCTTGTGGCTGTGAAGGCCCTGAAGGATCCCACCCTGGCTGCCCGGAAGGATTTCCAGAGGGAGGCCGAGCTGCTCACCAACCTGCAGCATGAGCACATTGTCAAGTTCTATGGAGTGTGCGGCGATGGGGACCCCCTCATCATGGTCTTTGAATACATGAAGCATGGAGACCTGAATAAGTTCCTCAGGGCCCATGGGCCAGATGCAATGATCCTTGTGGATGGACAGCCACGCCAGGCCAAGGGTGAGCTGGGGCTCTCCCAAATGCTCCACATTGCCAGTCAGATCGCCTCGGGTATGGTGTACCTGGCCTCCCAGCACTTTGTGCACCGAGACCTGGCCACCAGGAACTGCCTGGTTGGAGCGAATCTGCTAGTGAAGATTGGGGACTTCGGCATGTCCAGAGATGTCTACAGCACGGATTATTACAGGGTGGGAGGACACACCATGCTCCCCATTCACTGGATGCCTCCTGAAAGCATCATGTACCGGAAGTTCACTACAGAGAGTGATGTATGGAGCTTCGGGGTGATCCTCTGGGAGATCTTCACCTATGGAAAGCAGCCATGGTTCCAACTCTCAAACACGGAGGTCATTGAGTGCATTACCCAAGGTCGTGTTTTGGAGCGGCCCCGAGTCTGCCCCAAAGAGGTGTACGATGTCATGCTGGGGTGCTGGCAGAGGGAACCACAGCAGCGGTTGAACATCAAGGAGATCTACAAAATCCTCCATGCTTTGGGGAAGGCCACCCCAATCTACCTGGACATTCTTGGCTAGTGGTGGCTGGTGGTCATGAATTCATACTCTGTTGCCTCCTCTCTCCCTGCCTCACATCTCCCTTCCACCTCACAACTCCTTCCATCCTTGACTGAAGCGAACATCTTCATATAAACTCAAGTGCCTGCTACACATACAACACTGAAAAAAGGAAAAAAAAAGAAAGAAAAAAAAACCCTGTAAGGCAGTTTGGCAAATATATATATATATATATATATTTATATATCTAACTATCTATCAATCTATATCTACAGAGAGTTACCTTCTCTAGTACACAGAGAAGCTGCTGATACAGAAAACCACAAGACTTTAACAACTCAGAAACTCTAAAATATTAATAATACAAAGGAAAATTCCCTTTGACTTAAGCTGTGGCCCATGCTTCTAATGCTACGGCTCTTTGGAGCAGAAGACCTGGACCATGCAGAGAGACAATCTTTGGGATGAGAGCTCTGGGACGGGAGGAGTTAAGGTGGTGCTCAGTCGCTGCTGTGTGTGTCTGGTACCCCGGAAGCTCACCACAGGCACATGTGGGGACTGCATGGCTGTGCTCAGCAAAGAAATGATGCCTGAGCAGGGTCCTTGCCTCCACTTCTTCCCCATTTCCTCCACTTGAGGACAATGTTCTAATTTGTCCATTCTAAAAAGTGTAATCTTGATGCTTTTGGGAATCAATGATGGCACCTACGGGTAAACACAGAACAGACAACCCCACAACACAAAACCATGACAAGAGCTGACAGCATCCATCCTGGGTGTAAGCTTCTTGCTGGAGTTGCAAAGGATGTGTCTTTCTATTCTCAGAACTTAAAGAACTGGACTTTCTGGAGTAAAAGAACCACAGAAGAAAAAATAGCTGAAACCTGAACCCTGCCAAGACACTGCCACCTGTCTTTTCCATCATTGCAAAGATTCCTTGGCCTCACCTCCCACTTTCAGGCCCCTGGGAAGCTTAGGGGGTTAGGAAACAGGTCCCCATGTTATCTTTGAATGTAGACACAGCACGCTTTAGGGTTGCAATAGCAAGAGACTTGGTTGCTTGTAGGTCATTATAACGAATGCCCCCTGGTCCTCTCAGTTTCTCATCCTGCTTCCCTAGATCCTAGACCCCACTTCCTGGGTCTCTACTGAGTCCCAGGTAAACCCCACTGCCAAGGAAGGGAGCCAGGTCTAGTGAGAGGCTGCAGCAGTGAGTGTTTCAGGAAAGCAAGCAGTGGAGTGGGTGAGAGCCAGCAGGGGAGAATCTACTTGGCACCTGTGCCAGGCCCCTTGGCATTCTCCTCACGTTTCCAGCCCCTGGGAGGATTGATGCATCTGCCTTTGAGCTGTTGTGAAAACGCAGGGGCTGAGAAATCACTTTTGTGGGCTGGGGTAGGGGGAAGGGCTGAGGCTTCTCTGGGAGATGTCCAATGAGCGGCCAGTGCTGCAGGCTTGGAATCTTAATGAGGCCTGAGCCCTGACCGGAGAGAGGGAAGGAAACATCTGTGCTGGGGCCTGCTCCCTCTGCCCCAGCACTGGGGAATCACTGCCTTCCTGGCTGCTGCCACATACCACAGTGGGGCCGACCTCTGGGCCTCCCTGTCACATGAAACATGGAAGCCAACAGCCTGGCCCCACAGATCTCCTCTGCTGGAGAGGAGAGGATGTGTTCCTCCCAGGCCACGGGGCCCCTCTGCCTTGCCCCAAGCACAGAGCTACCTAAGGGACTGACCTTTGCCACACTGGAGCTCACCTCCCGGGTGCCTCTGGCCTACCCTTTTCCCTGCCAGCTGAAGAGACTGCCGGCAGCCTGGACGTCCTGGTTAGCTGAAGGCAGCTGAAATGTGGGCCTCCCTATGTGGGGTTTAGTACTGAATTATGTGTTTGTGGTTTTGCTTCTTTTTGTAGACCCACAAATATTTTTATTTGAATAGTAAAGAGCTGTGCTCAGATTGTAGCCATGGTTGGGATTTGGGATCAACGAGGCTGGTTAGCTGGACTGGGAGGGGAGGCAGGTGAGATGGGAATTTGGTGTTGGTTTTCTCACTGTGCTGTGCTCCTAGCCCCTGAGGACTTTGCAGGAAAGCTGTTTGGATTGACCACCTCAAGGAACTCCTTTGCTGTCCTATGACAATTACTCCTCTTGTCTTTCCACCTAGAGGACCGTTATGCCGGGGCTGTGAGTTCTGGCTGAAGCTGCACAATCTGGCTTTGATCTGGGTAGCTAAGAACAGGTGTTCAGGGAGGGCATGGTCTGGAGGCGCCTGAGGGAAAAGTATCTTCTGGTTAGAAGTGAAGCACCAATGCTCCTGTTGCTTCCCTTCTCCCATATCCTCTCCTGAACAAACACTGTGTGGCTTCTGTCTTCTTGGCTGTGTGTTTCAAAACTGATAGTCACTGATGGGCTGGGAAGAAGCAGGAGAAGAACTGGGTTGGAGATGGGTGGAAAACTGGGAATAAGGACCATGCTATGGTTTTATCTTCAGGGCACTGATTCATCATGGCCTAATGAAAGAAGGTGATTCCTTGGGGGAACAAGAAGCACTGAAGGAGACCGCAGTTGTCAGATTGCTGCCACCTTAGGCCCACGGGCTTTCACGCTGATCTTGGGGTCATGTTCTTACTGTGTCCATTGTAAGCAGCTGGCTGTGGAAGAGTGCCAGAGAGAGAGGGGCAGAGCAGGGAAGGGTAGCGAGGGAAGATATATTACTTTGATATATTAGCCTTGGCAGATTAAAAAAAAAAGATTTATTTTGGAGAGTAAAGGAGGCAGAAAATAGGGCCATGGAAAATGCAGGGTACACTATGGCTCTGGGGAGTGTGGCATGTACTGGGACACACCTGTCCCTCCTATATTGGGACAGGACTGATAAGGCTTAGTACACTGTATCTTAATTTCCCCATCTGCAGAATAAGGGGTACACCTGCTAATCTCTGGCCCCTTCTAGCTCTGAGTTCTTAGATTCTGACAACGTTGTGTCACTGTGCCATTTTTCTCAATACCATTCTGGAATGGCAGGACAGCCTTGCATAGGTGGGGGGCAGTTGCCGCTGAAGGCTCACTGGGCCTGGGCAAGGCTGAGTTCCAATAAGACACTCCTCTCCTTCTTTGCCCCTTGTCTCAACATTTCGGAGTCTGAAGTGTCTGAGACGACTGTAAGCCAGGAGGGGAGCATTTGGTCGGCTTCCCATCCTGGCATTCCTACATACCAGAAAAGCACTGTCGACGGTCCATTTATTTATTTATTTATGAAGAGCCTAATGTGATGAAATGTGTTGTGAAATGACATCCTATTAGCCAATATGGCCACTCCAGTTCATTTCACCCTCATTTTCCCATACCAAAGTCCACCTTTTAGAAGACCATGTCACATCGCCTCTGAGAGCTTAGCTTTGAAGTGAGCTCTTTTGTGTCTTTTGATGAACGCTCCAGGGCACCCAGGCTGGTCTCTCTGCCATGCCAACCTCACCGGATCCTCTCCTGGACTGAGTGAGAGTGACCTGCTGTTGGGTGTCCACCTTGGAGTAGGGTGTTGTGGCCATGGGACCTCCCAATTCTTCTTTCTTTGGATGTGAACCTAACTCTGCCCTCTAATGATATCCCTGAACTCTGAGGGGACCTTGGGCATCACTGAACCTCCTCTAGTCAAGAGACAAGAAAAAGGAGGTGCTGCTTCCTCCATTCAGCAGATCATGCACAAATTACTCCTAGACAGAGGCTTCCCAATTGAGGCAAAGGATCTTTGGTCCTTCTGGGTTCAGGTTGAACATTAATGTTCTTCTCTAGTGATGCCTACTTTGCGGAATACACTAGTGCAAGTCATTTTGGTGCTAAATACTGCAGAAACCAACACCAACGGGAGAGCTGTCAGCCAGCTTCTGGGGCTGCACGAGACTCAACTCAGAAGTAGAAGTGAGTCAGCCATGCCGGGAGAGTACAGGGTGCATTGGAAGTTAAGTAGAAAAGTGGCAGGGTGAAAAGACCAGCAAAATTTCTGATTTTGCTATTAGTTATCCACGTGGCCTTGGCCAAATCAGCTTATTTCTCATTTGAGACGTTTAAGAGGTTTGTATAGATATTTTTAGAGTCCTTTTCATTTTTTTAAAGCTAGGTTCTGGTCCCACCTGATATATATGTACTTGCTTGTTAAAAATAAGAGATGAAGTGAAAGATAAGGAAGGAGACGAAGAAAAGTTACCAATGGCAAAGGATTGATTATAGAAGACAATGAAAGAAGGCCTGGTGACTGGTAGATTTGAGGAATATTGTACAGGGAGCCAGGCAGACAAGCCAGAGACTTCATTTTTATGCTCTTCACAGGAGGTCACTGGCCTAGCCACTTGCCTGCGTTTCACCAGGCTGTTCCCCTCAGCTAGTCAACCTTCCCTGTTCAAAACCAACATGCTAACACTTGGCAGAAACTATTCCAAGATATGGAGAGAGGCCTGCCCATCTCTCATGCAGGCCACTGGAAATCAACTCTGGGTCAAAACCGACAAGATTTTTATCTTTTAATGCTTATTGTAAGAGGCCGGGATGTTTGAAAGAGGGCAGCGTCTTAAGTTGCAGACTATTCACCAAAGTCCTTTTCTATTCAGGAATGACTAATGACTTTCTTTGTTCCCCTTTTCCAAACCAAACAGCAGCAGTATTACCTTGACTCCAAAGTACAGTGATTGCAATGGACAGTCTTGAGATTAAGGGTCTTTTTAGTGCTTTGCTCGGAATCTGGAGCAAGGAAATGTCTAGGGATGTTTGTTGAATGAACCCACCTGCCAGCAGTCCAGAGAGCTGTCCCAAAGGCCCCTGAGATGTCTATTGTCTGCTTTGGTAGGGAGCCAACCTGCTAGAAGACTCAAGGGCATTTGCCTTAGAGAGAAAAAAAAATGGCACATTTAGTTCCGGGCTTGGGTGCTGAAGTCACTGTGTTTTTGGCTCCTGAAGCTCCACTTCCTTTTTTCATACCAAATAAATTGTTTTGGCCAATATTTAAGGATTTAAATATTTAAGGTTTTGAAGGAAAATTCCAAGTTTATGCATCATTATTAAGACATCAATATGGGCTTGGGAAAATGTTTTGGTGTTGCTTAATAATACTGGGAGTGTCTTCTAAAGAAAGTATACATATATGTACGTTGTGTGTTTATTTCTATATATTGGATTGGAATGGCATCAAAGAAGGCCTGTTTGAAGTCAGCAAATTGATTTCAATTTGGGATTCTCTTTTAGCCTTACCGTTCTTGGGGATAAGAAATCTGGAGATGAACTGGCCTCAGAAACATTCAGGAAGATTACGATGGGAAATCTGTCATCTTGTAAAGTTGGATCATTTGGTGCTCTCTGAACTAAATTATGCTGACTAACATTCTGAGCATCTGAAAATCATTCTTCCTGGTTGCCTGATAGATGACCACACAGGAGTCATTTCTATCACATCTTATTTGGGGGTTGGTATAGACGCCACCAGCACTGCAGGGAAAACCACCCCCTAGGAGGGGTGAGACAGAGAGAATGTGAGATTTTGATCATTGATTGCTCTTTCTGAAAGGCACTGCTGACCATAGTAGGTGTGGTGAATGTTTGTGTCACTGCATTGGGTATTAGAACTTTTGCTCTGAGCATGGGCTGCCCTACCCTGAAGAGCTGAATCTTGGCTTCTGACAGAAGCTCTGCATGACTCATTCTCCTCAAGCATATTTTCCTTATTTTCCTATGAAAGTATCTATAGTTTGCAACAAGAACTGTCTTTGGTTCAAGTGTCAGTGTACTAGAATAGATGCTTTTCTGTGAGACCTGTGGAGGTTGGATTGGGGTGGGGGGTGTTCCTTCAAAGTACTTGGAATTCTTTATCAAAGAAATAAAACTCAGATTCTCTTCTTTCTAGTTTACTTCTAAGTTCCAAATTTAAAACAAGACCCAGTTTGACATTTGCAGTAGGAGATAATGTCTTGAACCAGTGAGCTTGGCTATCTTTCTTTCATCATAGGGAGGCAGGGCAGTAAGTTGTATTAATTTTTCCTAATCACCCAGGCAAATATTGCTTTTTGTTCAGTGTCGTGACCCACATTCATAGCTTGCAGGGGATACTTCTAGCTGACAAAGCTGGAAGTTTCTAATAGAAGATGTATCAAGATAACAGAGGTGAGGAAAGGCCTGTTGAACATCTACTTCTGAGAATCATTGAGGCACTCAGGTCTTGCTTTTCAGTTTAGTATCTTACAAAAGGAGTTGGTTTCTTCTCATTTTAGCAATGATAAATTTAGACATCTTTTTTGAAAGGCCAAAGGTAGATTGAGAGCTCAAATACTTACGGTGTACTCTGACAGTCTTTTATTCTATTTTTCAAGACTTATTTTATATATACATATTTTTTATTTCCATAGGTTATTGGGGAACAGGTAGTGTTTGGTTACATGAGTAAGTTCTTTGGTGGTGATTTGTGAGATTTTGGCACACCCATCACCCAGGCAGTATGCACCATATTTGTAGTCTTTTATCCCTCACTCCCTTCCCACACTTTCCCCCTGAGTCCGCAGAGTCCATTGTGTCATTCTTATACCTTGACATCCTCACAGCTTAGCTCCCACATATCAGTGAGAACGTACGATGTTTGGTTTTCTATTCCTGAATTGCTTCACTTAGAATAGTGGTCTCCAATCTCACCCAGGCCTCTTCGAATGCCATTAATTCATTCCTTTTTATGGATGAGTAGTATTCCATCATACATATATATGTTTGTGTGTGTGTGTGTGTGTGTGTGTGTGGTGTGTGTGTGTGTGTGTGTGTGTGGGTGTGTGTGTGTGTGTGTGTGTGTGGGTGTGTGTGT
Immagine you found this fusion on your sample for the first time and you want to know if in other tumors are founded. What can you do?
Please go to this site:
and search information on the fusion.
## Hand On 4
Please evaluete FUS-DDIT3 from defuse results on Simulation 50X. Use IGV for see reads spanning or encomppassing.
cd ~/fusion/tutorial-fusion-transcript-2017/Results/Defuse/
``` then open IGV and look that fusion reads.
http://www.scienziofilia.altervista.org/dati/Chimeric_sort.bam
Write the name of the genes using a space and search the encompassing reads.
After that search information on this fusion on [cosmic][http://cancer.sanger.ac.uk/cosmic/fusion]
Well done!!!
